Project Description:
Raising public awareness of the risk of future pandemics due to antibiotic resistance has become a major objective of public health in recent years. This follows a decades-long deficit in discovering new antibiotics due in part to lack of investment but mainly to saturation of current approaches. To help mitigate both obstacles, Science à la Pelle (SciPelle) will enable direct contributions from the public to discover new antibiotics.
Project Type: Sustaining
Theme: Healthy Planet, Soil
Mentor: Jacqueline Goldin
Digging for Antibiotics
Currently, antimicrobial-resistant infections claim the lives of 1.27 million people annually.
The World Health Organization projects this grim toll to escalate to 10 million deaths per year, posing a substantial threat to global health. Consequently, the pursuit of new antibiotics and the elevation of public awareness regarding the risk of future pandemics due to antibiotic resistance have become paramount in public health initiatives.
Over the past half-century, the pace of discovering new antibiotics has slowed, and the emergence of resistance among pathogens is rendering existing antibiotics increasingly ineffective. Addressing this urgent need requires alternative approaches to antibiotic discovery. Recent advancements in microbial genome mining technologies provide a promising avenue to expedite this process.
Antibiotics are encoded in microbial DNA as Biosynthetic Gene Clusters (BGC), containing the necessary enzymes, transporters, and regulatory elements for biosynthesis. Modern DNA sequencing techniques enable the rapid assessment of bacterial strains to determine their potential to produce novel antibiotics.
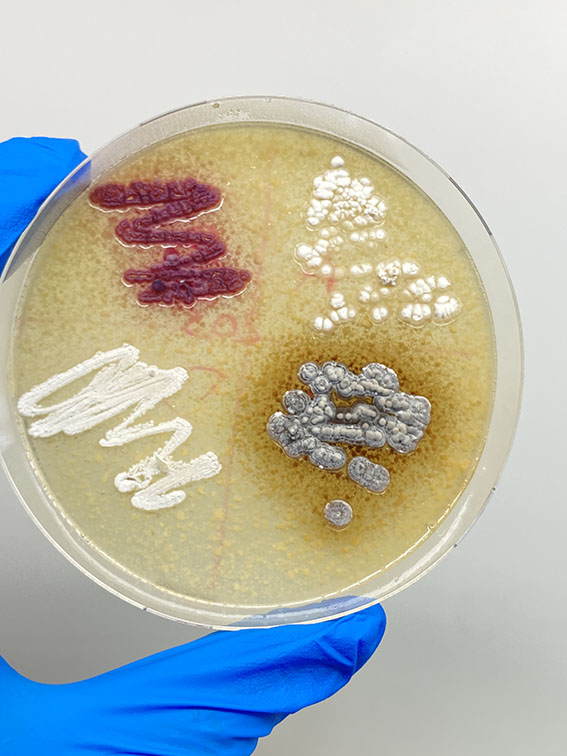
In response to this challenge, INSERM researcher Vincent Libis developed a systematic antibiotic discovery strategy, focusing on Actinobacteria, a bacterium prevalent in soils known for producing various antibiotics. His team aimed to establish a comprehensive collection of Actinobacteria to maximize the chances of discovering undiscovered BGCs that could combat antibiotic resistance.

To achieve this goal, the Center for Research & Interdisciplinarity (Inserm U1284) initiated a participatory science campaign, encouraging participants to collect soil samples during walks or hikes, record GPS coordinates, take photographs, and send the samples to the research team via a dedicated app named Science à la Pelle. This initiative aimed to create the largest and most diverse soil microbiome collection in France.
By February 2023, the team successfully developed a protocol to isolate bacteria from soil samples on a large scale, collecting approximately 2600 Actinomycetes from 500 Parisian soil samples. The sequencing of a batch of 1000 Parisian strains revealed 508 distinct unknown BGC families potentially encoding new antibiotics. However, the analysis suggested that the biosynthetic potential in Parisian parks would soon reach saturation at around 3000 strains.

To overcome this limitation, U1284 benefited from IMPETUS mentorship to develop SCIPELLE, a nationwide soil collection campaign during the summer, deploying a scientific mediation strategy, including a dedicated website, mobile app, and social media campaigns. The initiative garnered substantial public engagement, with over 1000 samples collected from 86 out of 95 counties in France.
The citizen participation campaign facilitated a high-throughput screening for new molecules produced by soil bacteria. The ongoing data analysis is broadcasted in real-time on an interactive map on the project website.
The success of the initiative is evident in the significant social media presence, press coverage, and engagement with citizens and educators, showcasing the potential of citizen initiatives to transcend the original scope of antibiotic discovery. As the project moves forward, the team aims to continue exploring new avenues in participatory soil research, building on the momentum generated by this impactful initiative.